
Insect Genomics and Bioinformatics. ZJU

Insect Genomics and Bioinformatics. ZJU

Published on: 2025年1月29日
Thanks to the fast development of sequencing techniques and bioinformatics tools, sequencing the genome of an insect species for specific research purposes has become an increasingly popular practice. Insect genomes not only provide sets of gene sequences but also represent a change in focus from reductionism to systemic biology in the field of entomology. Using insect genomes, researchers are able to identify and study the functions of all members of a gene family, pathway, or gene network associated with a trait of interest. Comparative genomics studies provide new insights into insect evolution, addressing long-lasting controversies in taxonomy. It is also now feasible to uncover the genetic basis of important traits by identifying variants using genome resequencing data of individual insects, followed by genome-wide association analysis. Here, we review the current progress in insect genome sequencing projects and the application of insect genomes in uncovering the phylogenetic relationships between insects and unraveling the mechanisms of important life-history traits. We also summarize the challenges in genome data sharing and possible solutions. Finally, we provide guidance for fully and deeply mining insect genome data.
Read more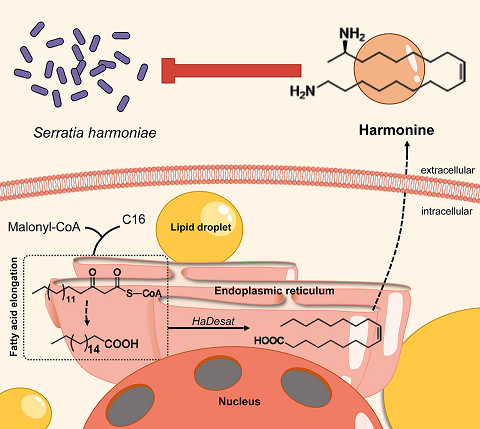
Published on: 2024年12月30日
Once prized for its use in biological pest control, the harlequin ladybird (Harmonia axyridis ) has become an invasive pest in nonnative regions, outcompeting local ladybird species. Here, we found that the harlequin ladybird safely harbors Serratia harmoniae , a highly pathogenic bacterium that causes severe mortality in other ladybird species. The harlequin ladybird’s tolerance to the pathogen is attributed to the defense alkaloid harmonine. Silencing three key genes in the harmonine biosynthesis pathway—Spidey , Sca2 , and Desat —reduced the production of harmonine, leading to increased bacterial levels and increased mortality. Penicillin treatment reversed this effect, reducing S. harmoniae content and increasing host survival. This symbiotic host–pathogen relationship confers an intraguild predation advantage to the harlequin ladybird.
Read more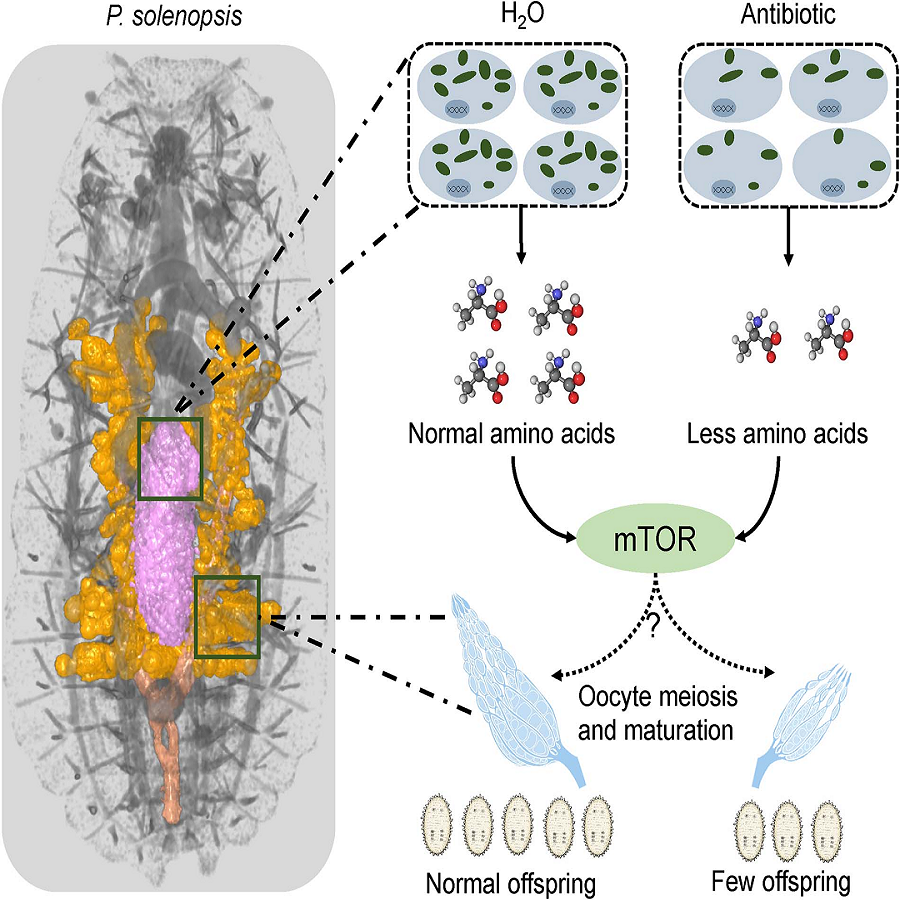
Published on: 2024年3月22日
The endosymbiont, “Candidatus Tremblaya phenacola” (T. phenacola PSOL), persisted throughout the complete life cycle of female hosts and was more active during oviposition, whereas there was a significant decline in abundance after pupation in males. A comprehensive analysis of amino acid metabolic pathways demonstrated complementarity between the host and endosymbiont metabolism. Elimination of T. phenacola PSOL through antibiotic treatment significantly decreased P. solenopsis fecundity. Weighted gene coexpression network analysis demonstrated a correlation between genes associated with essential amino acid synthesis and those associated with host meiosis and oocyte maturation. Moreover, altering endosymbiont abundance activated the host mechanistic target of rapamycin pathway, suggesting that changes in the amino acid abundance affected the host reproductive capabilities via this signal pathway. Taken together, these findings demonstrate a mechanism by which the endosymbiont T. phenacola PSOL contributed to high fecundity in P. solenopsis and provide new insights into nutritional compensation and coevolution of the endosymbiotic system.
Read more
Published on: 2023年4月3日
Here, we constructed a database by integrating the genomes and gene information of geminiviruses, their plant hosts, and insect vectors, named GPIBase (http://gpi.geminiviridae.com/). This database contains a complete set of nucleotide sequences from representative members of all 520 geminivirus species (one per species) and 119 betasatellites currently accepted by the International Committee on Taxonomy of Viruses, as well as 146 plant hosts infected by these viruses and six insect vectors. Interactions between plants, insects, and viruses are analyzed and presented, and functions of searching, genome browser, and phylogenetic analysis are also included. In general, GPIBase provides a widely accessible, simple, user-friendly, and knowledge-oriented platform. Users can retrieve information related to geminiviruses for studying tripartite connections between geminiviruses, plants, and insects
Read more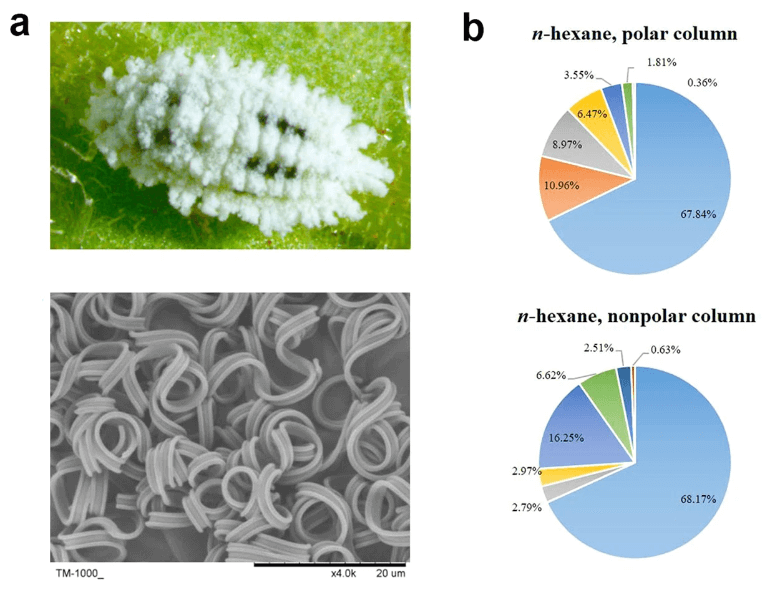
Published on: 2022年10月19日
Mealybugs are highly aggressive to a diversity of plants. The waxy layer covering the outermost part of the integument is an important protective defense of these pests. However, the molecular mechanisms underlying wax biosynthesis in mealybugs remain largely unknown. Here, we analyzed multi-omics data on wax biosynthesis by the cotton mealybug, Phenacoccus solenopsis Tinsley, and found that a fatty acyl-CoA reductase (PsFAR) gene, which was highly expressed in the fat bodies of female mealybugs, contributed to wax biosynthesis by regulating the production of the dominant chemical components of wax, cuticular hydrocarbons (CHCs). RNA interference (RNAi) against PsFAR by dsRNA microinjection and allowing mealybugs to feed on transgenic tobacco expressing target dsRNA resulted in a reduction of CHC contents in the waxy layer, and an increase in mealybug mortality under desiccation and deltamethrin treatments.
Read more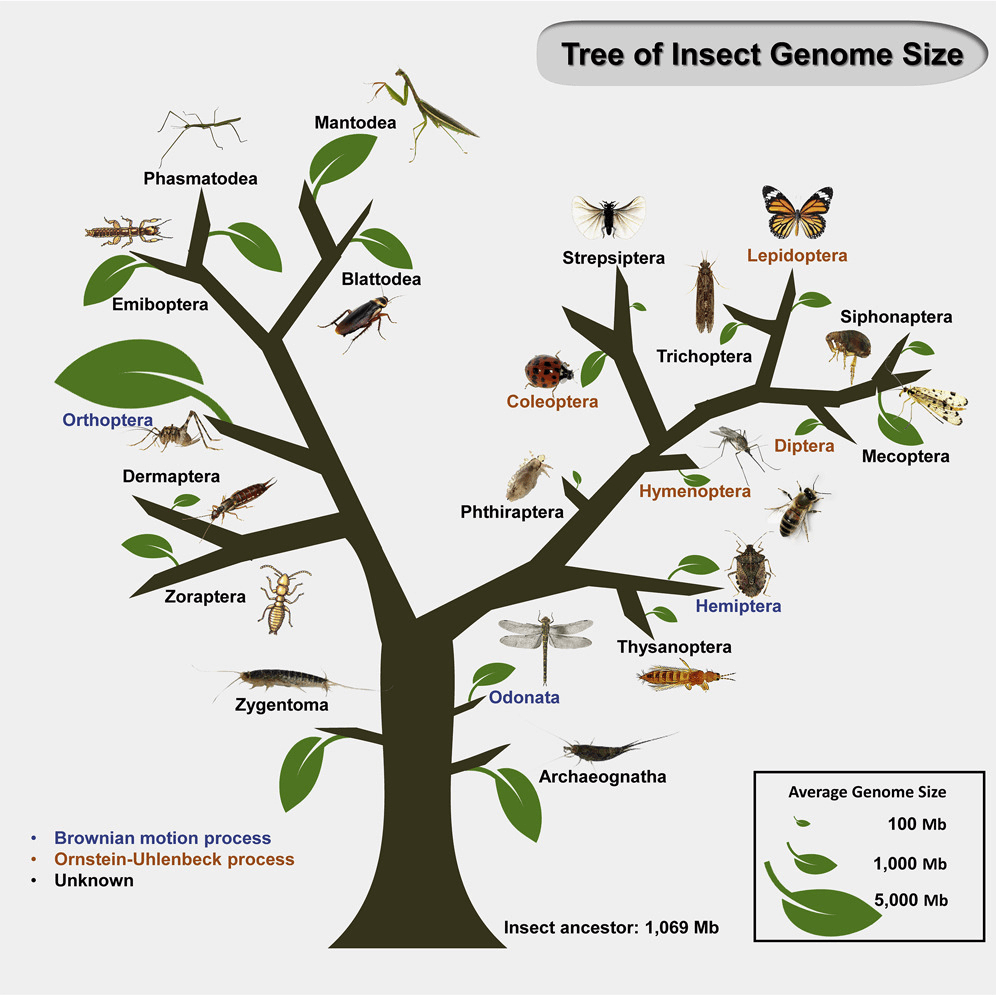
Published on: 2022年8月3日
Genome size (GS) can vary considerably between phylogenetically close species, but the landscape of GS changes in insects remain largely unclear. To better understand the specific evolutionary factors that determine GS in insects, we examined flow cytometry-based published GS data from 1,326 insect species, spanning 700 genera, 155 families, and 21 orders. Model fitting showed that GS generally followed an Ornstein–Uhlenbeck adaptive evolutionary model in Insecta overall. Ancestral reconstruction indicated a likely GS of 1,069 Mb, suggesting that most insect clades appeared to undergo massive genome expansions or contractions. Quantification of genomic components in 56 species from nine families in four insect orders revealed that the proliferation of transposable elements contributed to high variation in GS between close species, such as within Coleoptera. This study sheds lights on the pattern of GS variation in insects and provides a better understanding of insect GS evolution.
Read more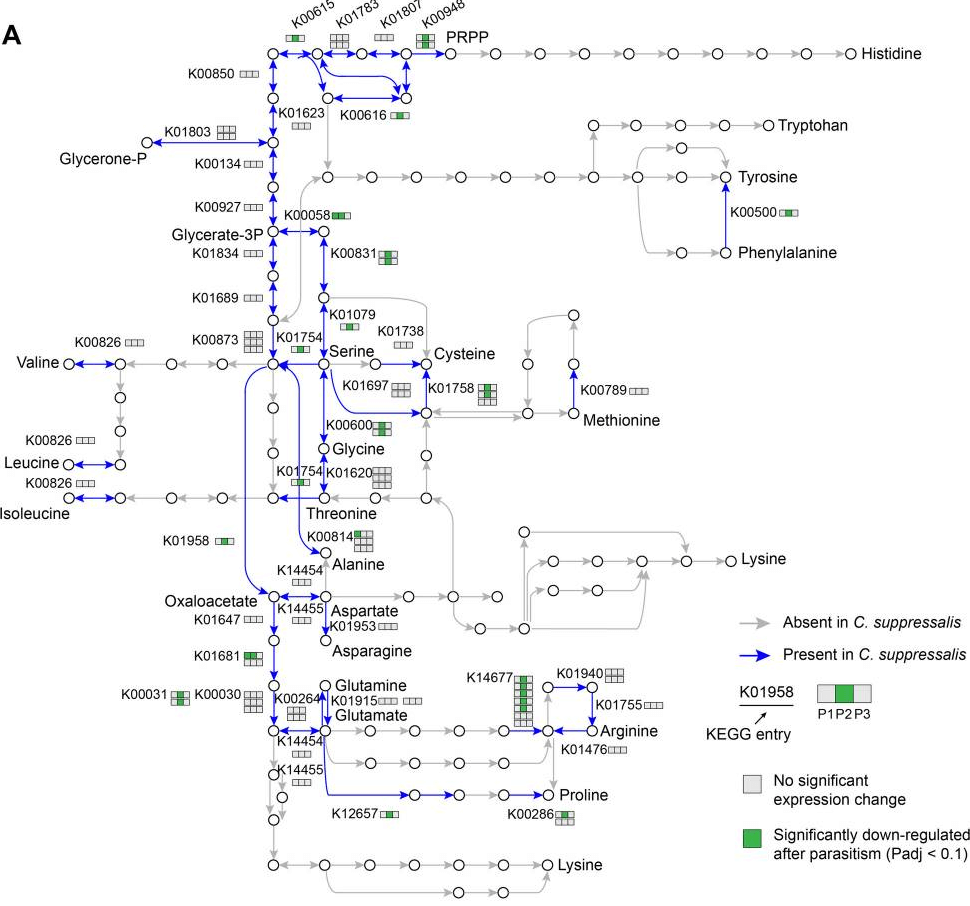
Published on: 2022年5月24日
This study delves into the nutritional exploitation of host organisms by parasitoid wasps, with a focus on amino acid resources. The genome of the parasitoid wasp Cotesia chilonis was sequenced, and its amino acid biosynthetic pathway was reconstructed. The analysis revealed that C. chilonis lost the ability to synthesize ten amino acids, including arginine. This loss of arginine synthesis was attributed to the absence of key genes in the arginine synthesis pathway. Transcriptome analysis suggested that parasitoid wasps inhibit amino acid utilization and activate protein degradation in the host. These findings shed light on the mechanisms of amino acid exploitation by parasitoid wasps and have implications for mass rearing of parasitoids for pest control.
Read more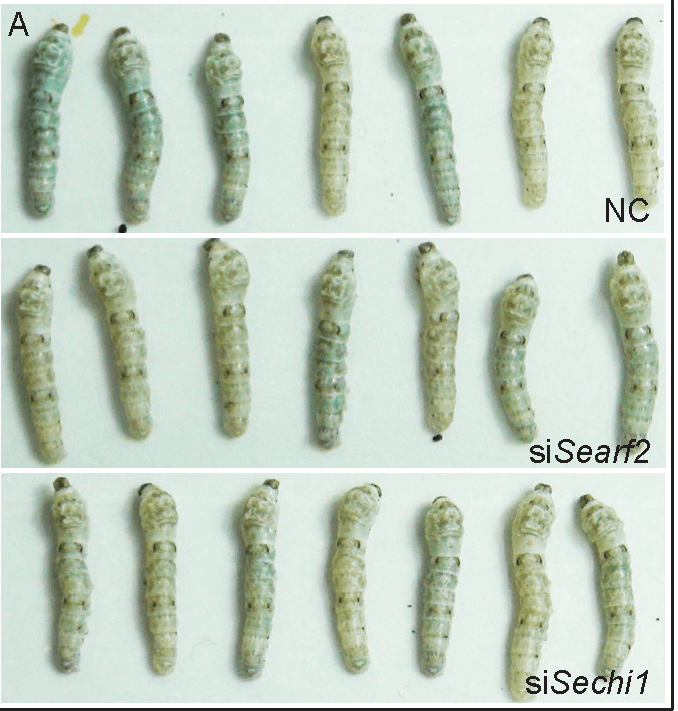
Published on: 2022年1月21日
This study investigates the off-target effects of RNA interference (RNAi) in pest control, focusing on two insecticidal small interfering RNAs (siRNAs) in both target and non-target insects. The findings indicate that off-target effects are widespread in both target and non-target insects. Gene expression changes were categorized based on homology to siRNA-targeted genes, related KEGG pathways, and continuous matches with siRNAs. The study also introduced the concept of Shannon entropy to assess the transcriptome profile balance after siRNA treatment, revealing that although hundreds of genes were affected, the transcriptome's overall integrity remained stable. This research provides evidence of siRNA cross-reactivity with individual genes in non-target species without disrupting the transcriptome profiles on a genomic scale. The proposed metric offers a means to estimate the off-target effects of insecticidal siRNAs, contributing to the safety assessment of RNAi-based pest control.
Read more
Published on: 2022年1月7日
We present an updated database, InsectBase 2.0 (http://v2.insect-genome.com/), covering 815 insect genomes, 25 805 transcriptomes and >16 million genes, including 15 045 111 coding sequences, 3 436 022 3'UTRs, 4 345 664 5'UTRs, 112 162 miRNAs and 1 293 430 lncRNAs. In addition, we used an in-house standard pipeline to annotate 1 434 653 genes belonging to 164 gene families; 215 986 potential horizontally transferred genes; and 419 KEGG pathways. InsectBase 2.0 serves as a valuable platform for entomologists and researchers in the related communities of animal evolution and invertebrate comparative genomics.
Read more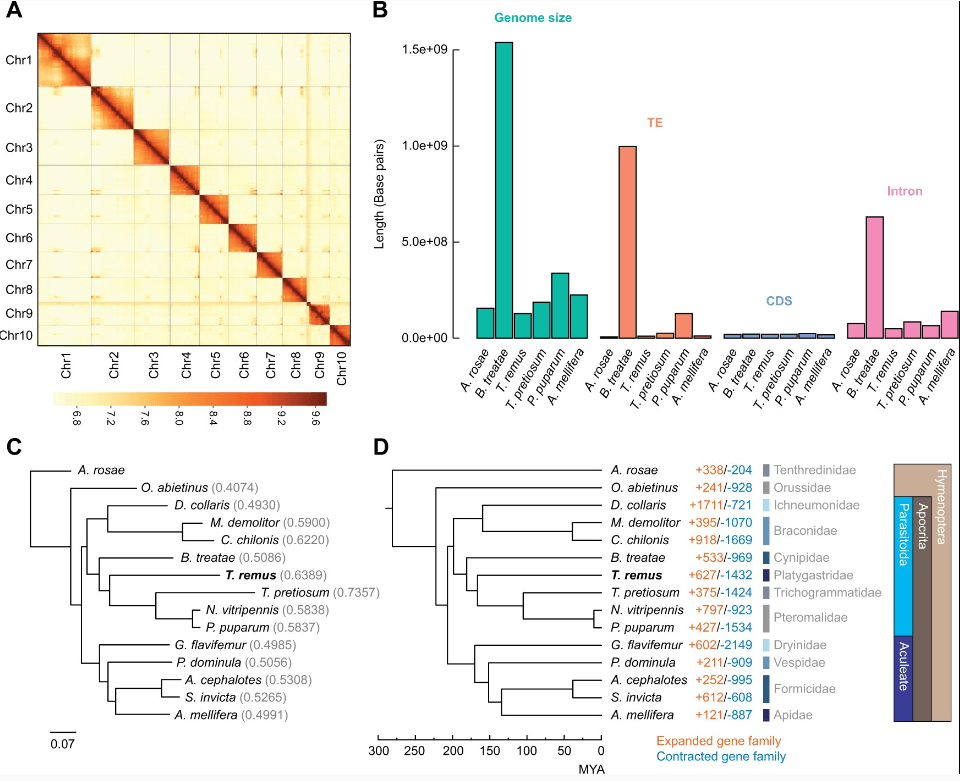
Published on: 2021年9月13日
Miniaturization, a widespread evolutionary trend in animals, has independently occurred in various parasitoid wasp lineages, contributing to species diversity. Telenomus remus, a small wasp species, has been studied to control the pest Spodoptera frugiperda. Its genome (129 Mb) has fewer repetitive sequences and shorter introns, resulting in a smaller size. Genomic analysis revealed accelerated evolution in hundreds of genes, particularly 38 genes related to eye and wing development, as well as cell size control, potentially influencing body size regulation. These findings shed light on the genetic basis of miniaturization in parasitoid wasps and provide insights into Hymenoptera evolution and pest control.
Read more
Published on: 2021年2月2日
A new high-quality chromosome-level genome assembly of the harlequin ladybird, Harmonia axyridis, was achieved in this study. This ladybird beetle is both a model organism for genetic research and a natural predator used in pest control. It became invasive in North America and Europe after being introduced for pest management. The assembly, totaling 423 Mb with an N50 scaffold size of 45.92 Mb, was generated by combining various sequencing technologies. Hi-C data allowed the anchoring of 1,897 scaffolds to eight chromosomes. Repeat sequences accounted for 51.2% of the genome, and after masking, 22,810 protein-encoding genes were annotated. The study also identified the X chromosome and Y-linked scaffolds.Additionally, a putative biosynthesis pathway for harmonine, a vital defense compound in H. axyridis, was established. This chromosome-level genome assembly is a valuable resource for exploring beetle biology and invasive biology.
Read more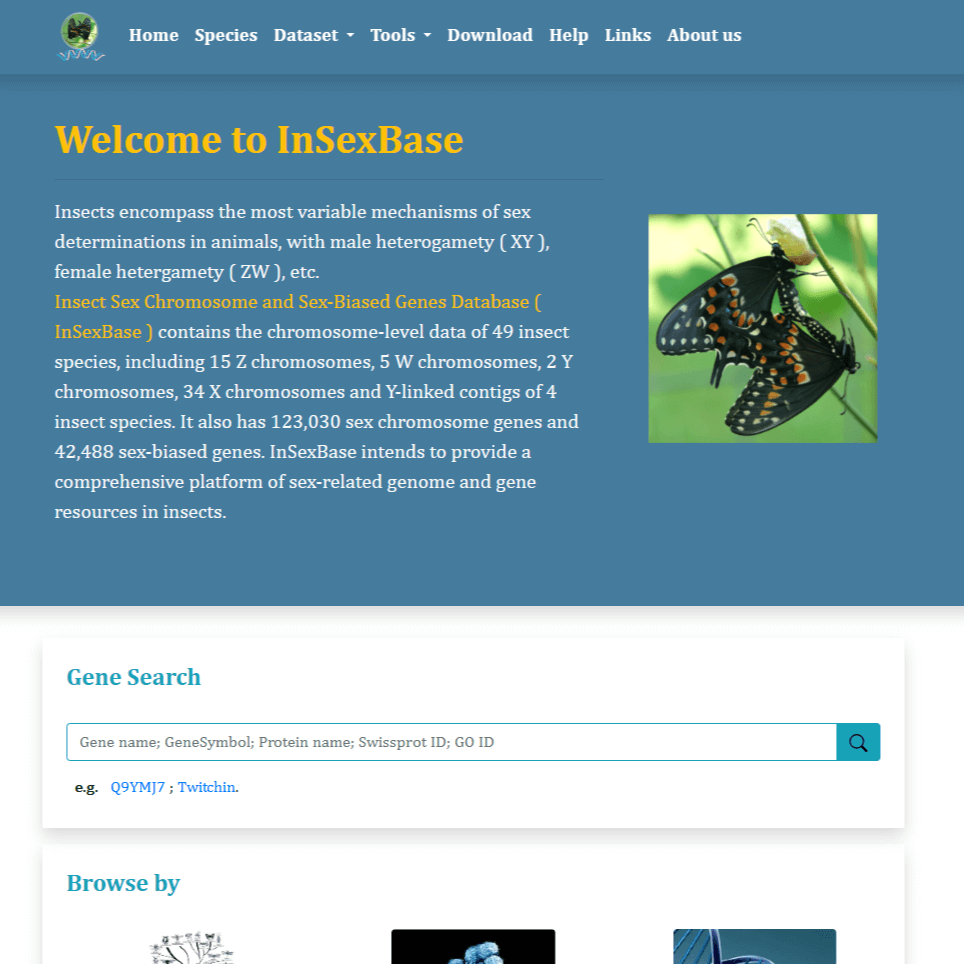
Published on: 2021年1月28日
Sex determination and the regulation of sexual dimorphism are key areas of interest in biology.Despite extensive research on sex chromosomes and sex-biased genes in numerous insect species, their gene sequences are scattered across different databases, often with limited annotations. In this study, chromosome-level sex chromosome data from 49 insect species, including X, Z, W, and Y chromosomes, were systematically collected. Y-linked contigs from four insect species were also obtained. These unannotated chromosome-level sex chromosomes were thoroughly annotated, resulting in a comprehensive dataset consisting of 123,030 protein-coding genes, 2,159,427 repeat sequences, 894 miRNAs, 1574 rRNAs, 5105 tRNAs, 395 snoRNAs, 54 snRNAs, and 5959 other ncRNAs. All these valuable data have been integrated into InSexBase, a user-friendly database dedicated to insect sex chromosomes and sex-biased genes.
Read more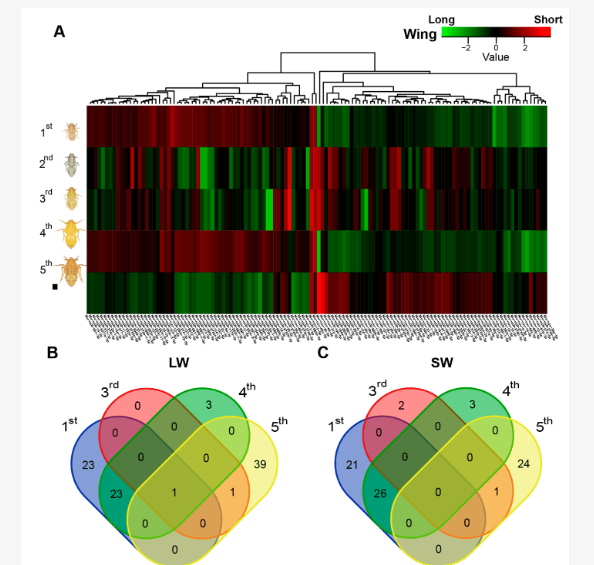
Published on: 2020年12月21日
Many insects exhibit wing polyphenism, developing different types of wings to adapt to various environments. The role of microRNAs (miRNAs) in regulating animal growth and development is well-established, but their involvement in wing polyphenism remains unclear. To explore this, we isolated small RNAs from different nymphal stages of long-wing (LW) and short-wing (SW) strains of the brown planthopper (BPH), Nilaparvata lugens. Sequencing revealed 158 conserved and 96 novel miRNAs, with 122 miRNAs differentially expressed between the two strains. Various miRNAs were upregulated in different instars of LW compared to SW, and vice versa. Predicted target genes were analyzed, implicating pathways such as insulin, MAPK, mTOR, FoxO, and thyroid hormone signaling, as well as thyroid hormone synthesis in wing form determination. Notably, miRNAs targeting genes in insulin signaling and insect hormone biosynthesis pathways were linked to wing dimorphism. Certain miRNAs, like Nlu-miR-14-3p, Nlu-miR-9a-5p, and Nlu-miR-315-5p, were confirmed to interact with insulin receptors (NlInRs). These findings provide insights into the miRNA-mediated regulation of wing polyphenism in BPHs, shedding light on how insects adapt to environmental cues through developmental plasticity.
Read more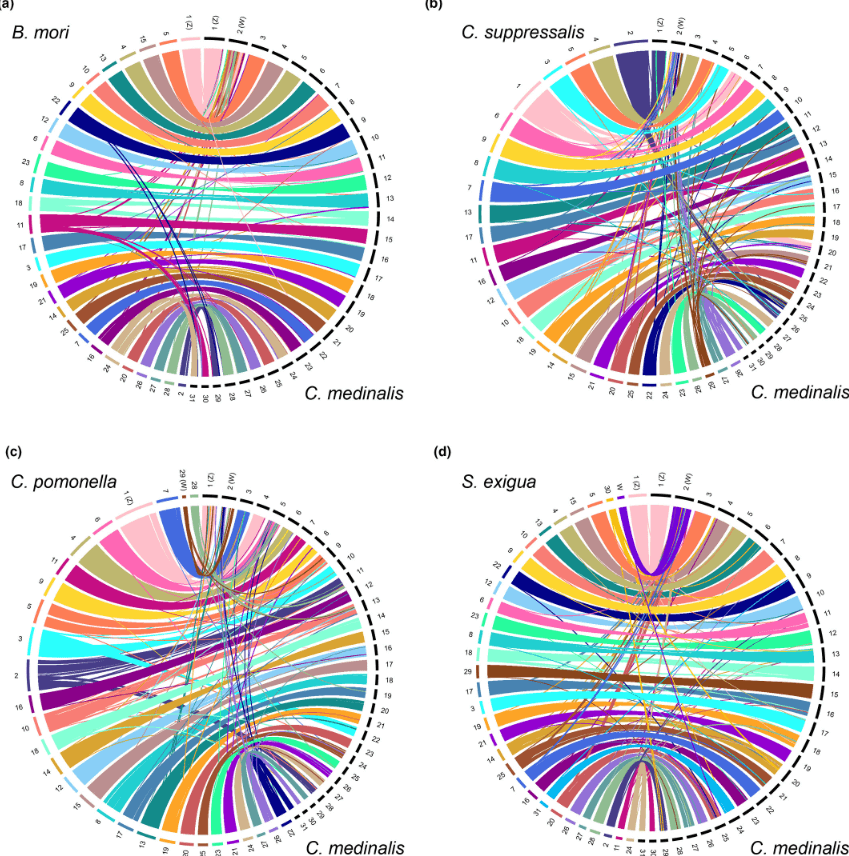
Published on: 2020年10月13日
The rice leaffolder, a destructive pest affecting rice crops in humid regions, had its genome sequenced using Illumina and PacBio technologies. This yielded a high-quality assembly of 528.3 Mb with 96.4% of BUSCOs detected, indicating completeness. The genome contains 39.5% repeat sequences and 15,045 protein-coding genes. Comparative analysis revealed expanded gene families related to hormone biosynthesis. Hi-C technology provided a chromosome-level assembly, showing chromosomal synteny with other lepidopteran insects. Z and W chromosomes were identified, with the W chromosome being notably complete at 20.75 Mb. This comprehensive genome assembly is a valuable resource for research on insect migration, chromosome evolution, and pest control.
Read more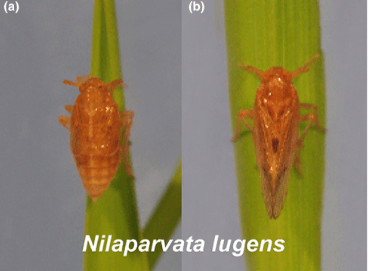
Published on: 2020年8月11日
The brown planthopper (Nilaparvata lugens), white-backed planthopper (Sogatella furcifera), and small brown planthopper (Laodelphax striatellus) are significant rice pests with genetic proximity but ecological differences. Although draft genomes of these insects exist, they require improvement. In this study, a higher-quality genome assembly of N. lugens was created using PacBio and Illumina platforms, with a contig N50 of 589.46 Kb. Chromosome-level scaffold assemblies for all three species were produced using HiC scaffolding, greatly enhancing scaffold N50s (N. lugens: 77.63 Mb, S. furcifera: 43.36 Mb, L. striatellus: 29.24 Mb). Sex chromosomes were identified through genome re-sequencing, revealing diverse gene content and high synteny. These chromosome-level genome assemblies serve as valuable resources for molecular ecology research and the development of pest control strategies.
Read more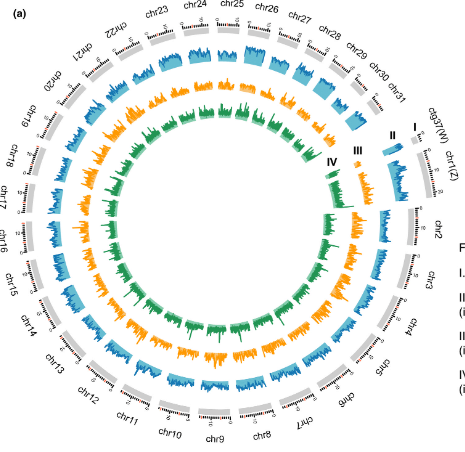
Published on: 2020年5月2日
The fall armyworm, scientifically known as Spodoptera frugiperda, is a destructive lepidopteran insect pest that inflicts significant economic losses. Over recent years, it has rapidly spread worldwide, but the mechanisms behind its swift dispersal have remained unclear. To address this, we present a high-quality genome assembly of the fall armyworm, referred to as the ZJ-version. This genome was generated using advanced technologies such as PacBio and Hi-C, with the sample collected from Zhejiang province, China, exhibiting high heterozygosity. The ZJ-version has a genome size of 486 Mb, comprising 361 contigs with an N50 of 1.13 Mb. Further Hi-C scaffolding led to the assembly of 31 chromosomes and a portion of the W chromosome, achieving a chromosome-level genome with a scaffold N50 of 16.3 Mb. Identification of sex chromosomes was achieved through genome resequencing of a single male and female pupa.
Read more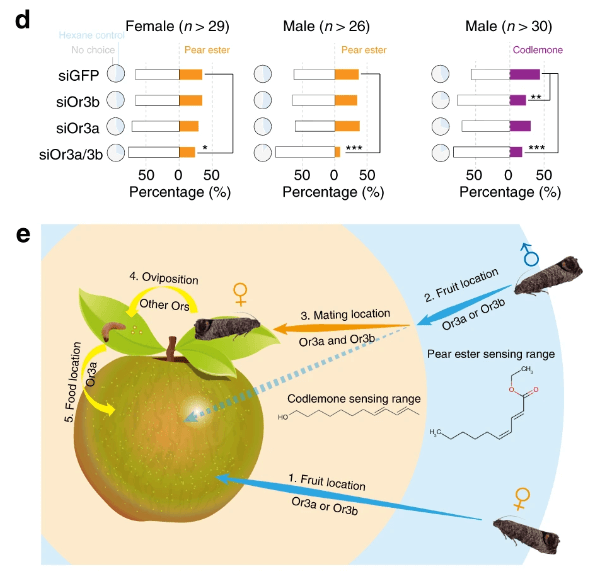
Published on: 2019年9月17日
The codling moth Cydia pomonella, a major invasive pest of pome fruit, has spread around the globe in the last half century. We generated a chromosome-level scaffold assembly including the Z chromosome and a portion of the W chromosome. This assembly reveals the duplication of an olfactory receptor gene (OR3), which we demonstrate enhances the ability of C. pomonella to exploit kairomones and pheromones in locating both host plants and mates. Genome-wide association studies contrasting insecticide-resistant and susceptible strains identify hundreds of single nucleotide polymorphisms (SNPs) potentially associated with insecticide resistance, including three SNPs found in the promoter of CYP6B2. RNAi knockdown of CYP6B2 increases C. pomonella sensitivity to two insecticides, deltamethrin and azinphos methyl. The high-quality genome assembly of C. pomonella informs the genetic basis of its invasiveness, suggesting the codling moth has distinctive capabilities and adaptive potential that may explain its worldwide expansion.
Read more